
Professor
Department of Biochemistry
Role of 3D genome architecture in health and disease
McIntyre Medical Sciences Building
3655 promenade Sir-William-Osler
Office: Room 815A; Lab: Room 815
Montreal, Quebec H3G 1Y6
Tel: 514-398-4975; Lab: 514-398-1070
Fax: 514-398-2603
josee.dostie [at] mcgill.ca
Dostie Lab website
Dostie Lab link to the Rosalind and Morris Goodman Cancer Research Centre: (Coming soon)
2000 - PhD, McGill University
| Accepting graduate students |
Research Interests
Background:
The human genome consists of approximately three billion nucleotides distributed amongst twenty-two autosomes and one sex chromosome. Most of our genes are encoded within these 23 chromosomes. With few exceptions, human cells contain two genome copies that if joined would measure over two meters in length. Chromosomes are thus extensively folded in order to fit within the ultra small, micrometer-scale confines of the cell nucleus.
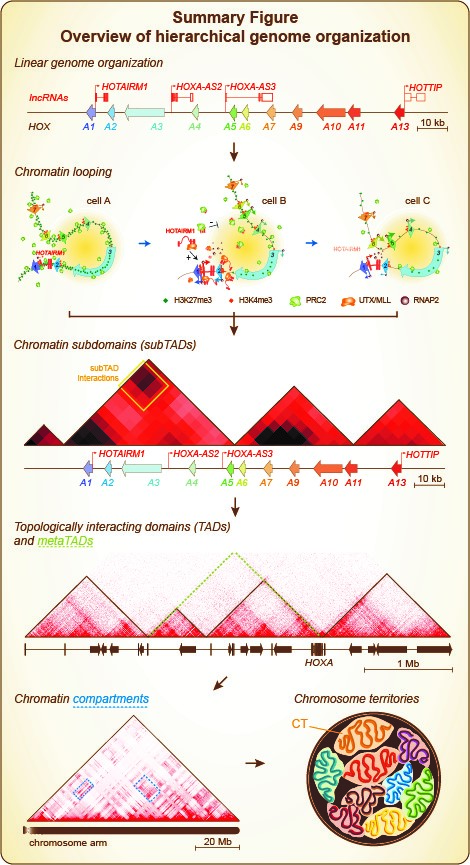
Our genomic DNA is first packaged with histone and non-histone proteins in the form of chromatin. This packaging shortens chromosome length at least sevenfold. Chromatin then folds different ways in the nucleus: from high to low resolution, chromatin fibers first engage into so-called “looping contacts” where distal genomic regions are physically close to each other (see Summary Figure)[1]. Looping interactions can create microenvironments and result in the formation of small chromatin domains or “subTADs”. These domains are specific to cell types, and vary mostly based on chromatin composition and whether genes are expressed. SubTADs can interact with one another within the larger “topologically associating domains” (TADs). These are more conserved between cell types and even across species at synthenic regions. TADs then interact with each other, forming “metaTADs” along chromosomes. MetaTAD formation broadly depends on chromatin composition and transcription activity. Large chromatin domains also tend to congregate as “compartments” in the nuclear space when they have similar composition and activity. Consequently to this extensive chromatin folding, chromosomes each occupy their own nuclear territory (CT), although chromatin from different chromosomes can also interact.
Research Overview:
The Dostie lab is interested in the role of spatial genome organization in human health and disease. Spatial genome organization refers to where chromosomes localize in the nuclear space and to how chromatin folds in three-dimensions (3D). We combine cell and molecular biology, genomics, and computational biology to understand how spatial genome organization impinges on the regulation of gene expression during normal cell differentiation and in the context of malignancies.
We use cells grown and manipulated in culture (gene inductions, RNA interference, CRISPR/Cas9) as our model systems. We use fluorescence microscopy (immunofluorescence, fluorescence in situ hybridization) to study chromatin localization. We apply classical molecular biology techniques such as cloning, real-time quantitative PCR, northern and western blotting, RNA and protein immunoprecipitation to identify and study underlying molecular mechanisms that regulate spatial chromatin organization and gene expression. We use both classical genomics (Chromatin immunoprecipitation, RNA-seq) and cutting-edge 3D genomics approaches (Hi-C, 5C) to map chromatin composition (epigenomics), activity, and folding. In collaboration with other groups, we integrate our findings computationally to uncover when, how, and why spatial genome organization affects the expression of genes (see Overview Figure).

I. Discovering underlying principles of chromatin organization.
We are working towards explaining why looping and chromatin domains form as they do and what makes them change. To this end, we are mapping 3D genome organization (Hi-C, 5C) under various conditions and relating this information to chromatin composition (ChIP-qPCR, 2C-ChIP, ChIP-seq) and activity (RT-qPCR, RNA-seq, CAGE). For example, as part of an ongoing collaboration we previously examined how chromatin folding varies during limb formation, and found that long-range contacts requiring the function of PRC2 (Polycomb repressive complex 2) also promote enhancer-promoter interactions [2]. In the past, we also tracked chromosome organization during cellular differentiation, and found that neighboring chromatin domains tend to naturally pair into progressively larger architectures (metaTADs) based on chromatin composition and activity [3]. Additionally, we previously compared data from microscopy-based approaches (FISH) to those from molecular biology-driven techniques (5C, Hi-C), and found that molecular-based techniques seem to capture more than the physical distance between chromatin segments [4].
II. Identifying novel 3D chromatin control mechanisms that regulate gene expression.
We are interested in identifying molecular mechanisms that control transcription through changes in 3D chromatin organization. We are currently studying how long non-coding RNAs (lncRNAs) alter the composition, 3D organization and activity of chromatin. LncRNAs belong to a large class of transcripts longer than 200 nucleotides that lack protein-coding sequence. Like mRNAs, they are transcribed by RNA polymerase II but tend to exist at lower levels and are more cell type-specific. Generally, lncRNAs are rapidly evolving genes with poorly conserved sequences. Unlike most lncRNAs, the mammalian HOTAIRM1 transcript can be identified by comparative genomic analysis and appears to have emerged prior to marsupial-eutherian divergence up to 160 Ma ago [5]. This lncRNA is thus expected to have evolutionarily conserved roles across all mammals, and interestingly, HOTAIRM1 has been linked to many cancer types.
We previously found that HOTAIRM1 is required to physically uncouple distal genes at the HOXA gene cluster so as to prevent premature transcription activating during neuronal differentiation induced with retinoic acid (RA) [6]. We also found that another lncRNA named HOXA-AS2 located downstream from HOTAIRM1, which is normally activated later during neuronal differentiation appears important to maintain high transcription activity at the HOXA cluster [7].
III. Genome organization in health and disease.
We aim to understand the role of 3D chromatin organization in cancer and determine whether it can be used as a molecular biomarker in the clinic. Proper genome organization is important for human health as demonstrated in cancers where SNPs (single nucleotide polymorphisms) creating novel enhancers activate distal genes long-range through DNA looping [8-10]. Accordingly, key developmental genes and those controlled by very strong enhancers often localize within stable TADs where regulation can occur independently from their neighbors [11]. Notably, the boundaries of neighborhoods wherein proto-oncogenes reside are sequences often mutated [12] or deleted in cancer cells, and their perturbation activates oncogene expression in non-cancerous cells [13]. In fact, actual TAD boundary disruptions can cause domain rearrangements, inappropriate gene activation, leading to carcinogenesis or other diseases [14].
We use both cell lines and patient-derived xenografts (PDXs) to map chromatin organization in normal and cancer cells with either Hi-C (genome-wide) or the 5C (genome-scale) technology. We then apply machine learning artificial intelligence (AI) programs to extract 3D chromatin signatures (3DCSs), integrate this information with other “omics” data, and identify potential molecular mechanisms and/or pathways deregulated in the corresponding cancer types. We previously examined the value of 3DCSs in disease classification, and demonstrated that it contains the necessary information to classify leukemia subtypes [15].
References
1. Fraser, J., et al., An Overview of Genome Organization and How We Got There: from FISH to Hi-C. Microbiol Mol Biol Rev, 2015. 79(3): p. 347-72.
2. Gentile, C., et al., PRC2-Associated Chromatin Contacts in the Developing Limb Reveal a Possible Mechanism for the Atypical Role of PRC2 in HoxA Gene Expression. Dev Cell, 2019. 50(2): p. 184-196 e4.
3. Fraser, J., et al., Hierarchical folding and reorganization of chromosomes are linked to transcriptional changes in cellular differentiation. Mol Syst Biol, 2015. 11(12): p. 852.
4. Williamson, I., et al., Spatial genome organization: contrasting views from chromosome conformation capture and fluorescence in situ hybridization. Genes Dev, 2014. 28(24): p. 2778-91.
5. Yu, H., et al., Evolution of coding and non-coding genes in HOX clusters of a marsupial. BMC Genomics, 2012. 13: p. 251.
6. Wang, X.Q. and J. Dostie, Reciprocal regulation of chromatin state and architecture by HOTAIRM1 contributes to temporal collinear HOXA gene activation. Nucleic Acids Res, 2017. 45(3): p. 1091-1104.
7. Wang, X.Q.D., et al., 2C-ChIP: measuring chromatin immunoprecipitation signal from defined genomic regions with deep sequencing. BMC Genomics, 2019. 20(1): p. 162.
8. Pomerantz, M.M., et al., The 8q24 cancer risk variant rs6983267 shows long-range interaction with MYC in colorectal cancer. Nat Genet, 2009. 41(8): p. 882-4.
9. Pittman, A.M., et al., Allelic variation at the 8q23.3 colorectal cancer risk locus functions as a cis-acting regulator of EIF3H. PLoS Genet, 2010. 6(9).
10. Meyer, K.B., et al., A functional variant at a prostate cancer predisposition locus at 8q24 is associated with PVT1 expression. PLoS Genet, 2011. 7(7): p. e1002165.
11. Dowen, J.M., et al., Control of cell identity genes occurs in insulated neighborhoods in mammalian chromosomes. Cell, 2014. 159(2): p. 374-87.
12. Ji, X., et al., 3D Chromosome Regulatory Landscape of Human Pluripotent Cells. Cell Stem Cell, 2016. 18(2): p. 262-75.
13. Hnisz, D., et al., Activation of proto-oncogenes by disruption of chromosome neighborhoods. Science, 2016. 351(6280): p. 1454-8.
14. Valton, A.L. and J. Dekker, TAD disruption as oncogenic driver. Curr Opin Genet Dev, 2016. 36: p. 34-40.
15. Rousseau, M., et al., Classifying leukemia types with chromatin conformation data. Genome Biol, 2014. 15(4): p. R60.
Publications (complete list) - Josée Dostie