We’ve moved
The Rosalind and Morris Goodman Cancer Institute can now be found at www.goodmancancer.ca
The Rosalind and Morris Goodman Cancer Institute can now be found at www.goodmancancer.ca
Professor/Professeur
Rosalind and Morris Goodman Cancer Institute
Department of Medicine
Faculty of Medicine and Health Sciences
McGill University

1. Harold E. Johns Award from National Cancer Institute of Canada, 2002
Dr. Yang’s laboratory focuses on the molecular and epigenetic basis of cancer, stem cells and animal development, especially about how cell signaling regulates chromatin modification, gene expression, and other events in the nucleus. His research program is mainly divided into the following three parts:
1) Characterization of two paralogous histone acetyltransferases—The gene for human MOZ (monocytic leukemia zinc finger protein) was identified in 1996 as a fusion partner rearranged in t(8;16)(p11;p13), a recurrent translocation associated with the M4/M5 subtypes of acute myeloid leukemia. Since then, this gene has been found to be a partner in three other chromosomal translocations linked to acute myeloid leukemia and therapy-related myelodysplastic syndromes.
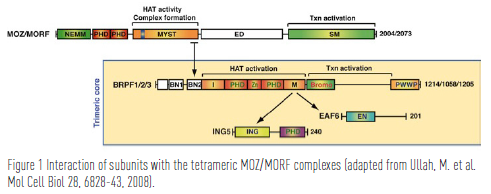
In 1999, Dr. Yang’s group identified MORF (MOZ-related factor) as a MOZ paraglog and discovered that MOZ and MORF are histone acetyltransfrerases. Like the MOZ gene in t(8;16), the MORF gene is rearranged in leukemia patients with t(10;16)(q22;p13). These two genes are important for different developmental programs in mammals such as maintenance of hematopoietic stem cells and neurogenesis. MOZ and MORF are catalytic subunits of multiprotein complexes containing ING5 (inhibitor of growth 5), EAF6 (homolog of Esa1- associated factor 6), and BRPF1/2/3 (bromodomain-PHD finger protein 1, -2, or -3). Importantly, these complexes are also formed by at least by one fusion protein from a leukemia-associated chromosome translocation. Dr. Yang’s group has recently analyzed how subunit interaction regulates the enzymatic activity of normal and leukemic MOZ and MORF proteins.
2) Function and regulation of histone deacetylases—Histone deacetylases (HDACs) are relatively novel enzymes that are able to remove acetyl groups from acetylated forms of histones and many other protein substrates. In the mammalian system, eighteen proteins have been shown to possess such enzymatic activity, with four of them (HDAC4, 5, 7 and 9) forming a subgroup known as class IIa.
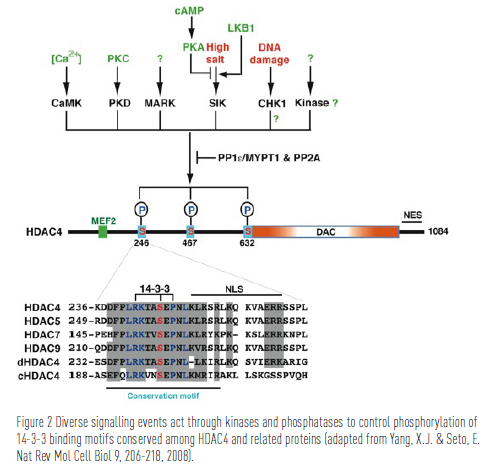
In 1999, Dr. Yang’s group identified HDAC4 and showed that this deacetylase interacts with the MEF2 (myocyte enhancer factor 2) family of transcriptional regulators to repress transcription (the process of making messenger RNA from genes). This repression is tightly controlled in cells: signal-dependent protein kinases phosphorylate class IIa HDACs, consequently driving nuclear export of these enzymes, and promoting MEF2- dependent transcription. This dynamic interplay is essential in different tissues as the MEF2 family plays a crucial role in a number of cellular programs, including muscle and bone cell differentiation, neuronal survival, programmed T cell death, and endothelial integrity. There are four MEF2 isoforms in humans, MEF2A, B, C and D. Dr. Yang’s group has been investigating how cell signaling controls the action of HDAC4 and its paralogs in terms of MEF2 regulation.
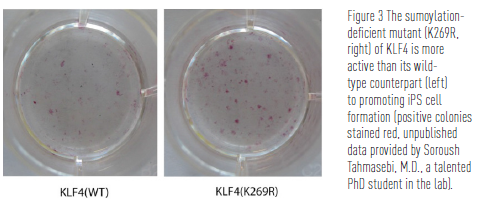
3) Post-translational modification in cell differentiation and reprogramming—By analogy to numerous modifications present in histones and the tumour suppressor p53, Yang has recently postulated that this also occurs in other major protein regulators. By use of MEF2 as an example, his group found a phosphorylation-dependent sumoylation motif conserved in MEF2 protein from worms to humans. A similar motif is present in the stem cell reprogramming factors of KLF4 and Sox2, so Dr. Yang’s group has been assessing the importance of such a motif in reprogramming fibroblasts into induced pluripotent stem (iPS) cells.